Coronavirus identification protocol in SciNote
![]() 3 min read
3 min read
In the light of the current outbreak of a novel coronavirus (COVID-19) many new challenges have stumbled in front of all of us. In SciNote we are well aware of the importance of the research community to join forces and contribute to the efforts to combat the challenges that the new virus has faced us with.
We set up an example of the experiment in SciNote for the identification of 2019-Novel Coronavirus by real-time RT PCR that has been published by the Centers for Disease Control and Prevention (CDC) (accessible here). We show you how to easily create a well-structured experiment in SciNote in only 8 simple steps:
CREATING THE PROJECT, EXPERIMENT AND TASK WORKFLOW IN SciNote
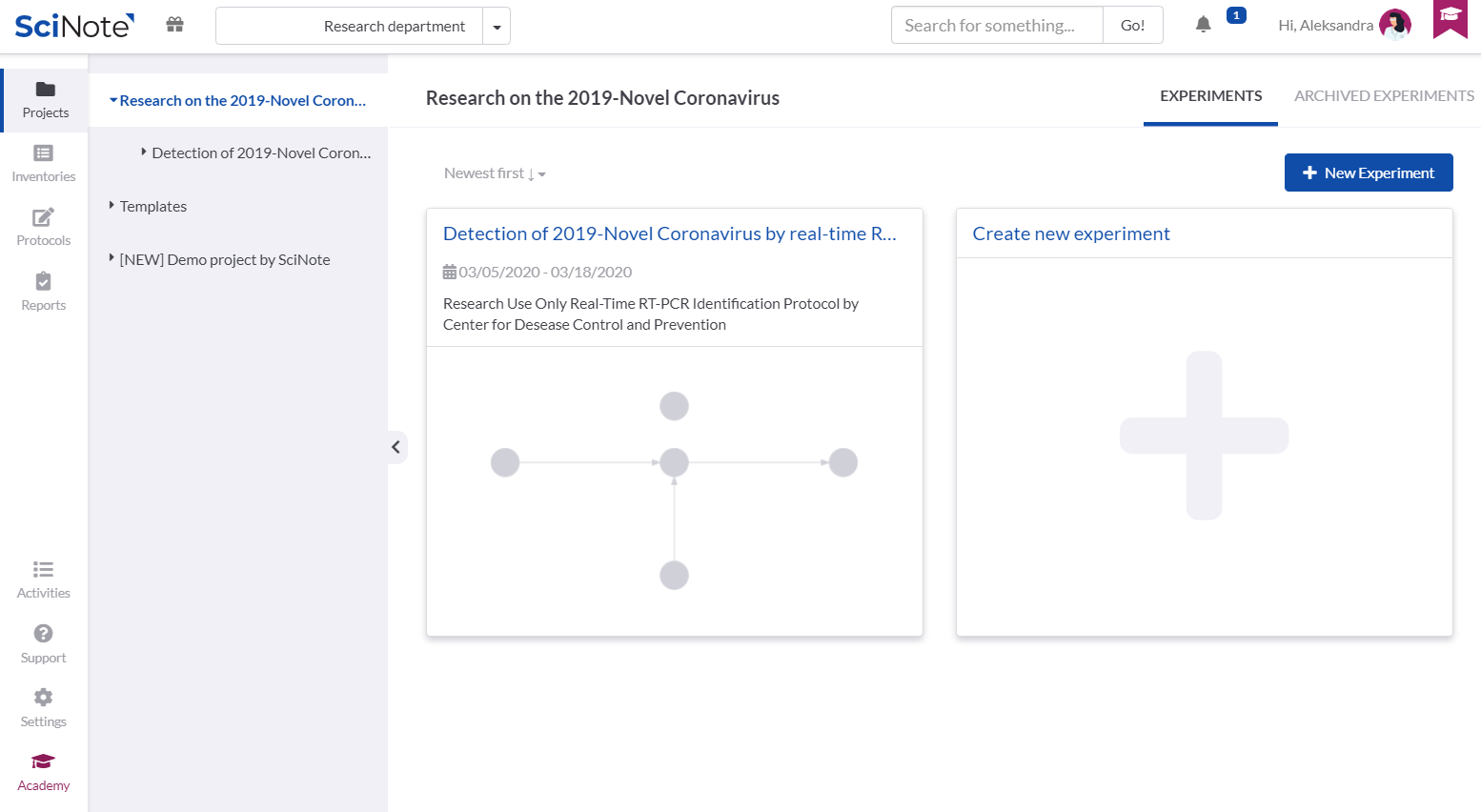
In the Project named Research on the 2019-Novel Coronavirus, we created the Experiment: Detection of 2019-Novel Coronavirus by real-time RT PCR.
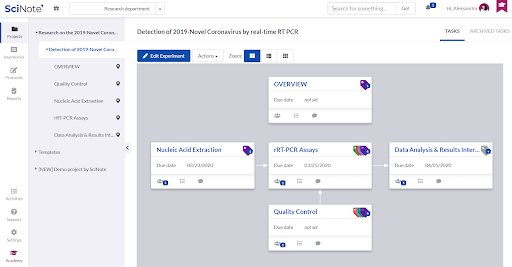
Inside the Experiment we created separated tasks that are required to execute the entire protocol. The tasks are connected with the arrows to visualize the required workflow. For each task we set up the due date and assigned it to the person that is best qualified to execute it.
ADDING TAGS ON THE TASKS
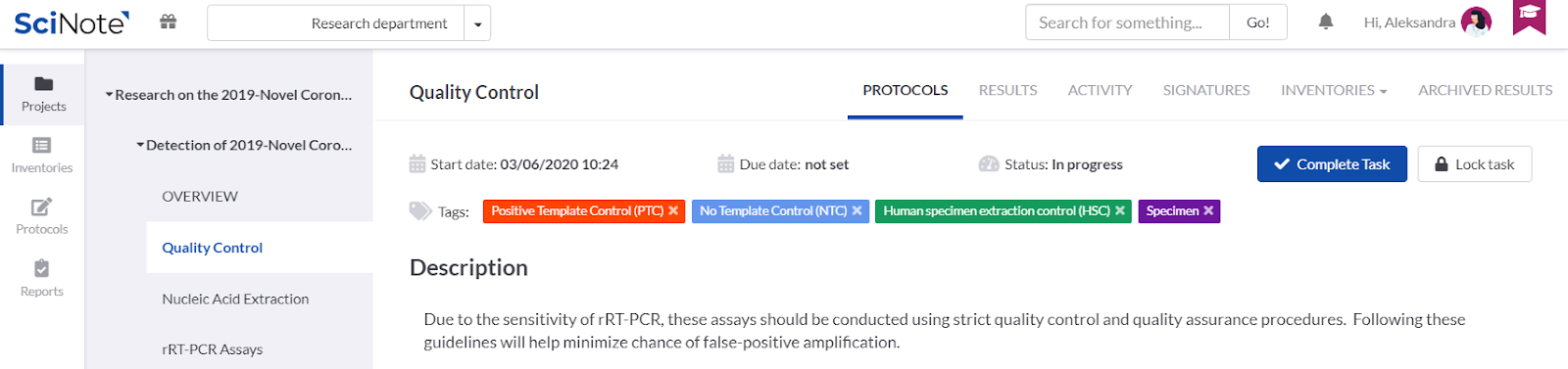
We added colourful tags onto the tasks to emphasize the control samples that have to be considered for the quality control on each stage of the process workflow. We could also use the tags for anything else we find important to visually emphasise, e.g. to label in which laboratory a particular task should be executed, etc.
IMPORTING THE PROTOCOL FROM protocols.io INTO SciNote
For the Nucleic Acid Extraction task, the referred CDC protocol lists several commercially available RNA extraction procedures as suitable to use. We therefore imported one of the external protocols following the recommended manufacturer’s procedure from the protocols.io platform (in the Protocols section, selecting External protocols tab) into the team’s Protocols repository.
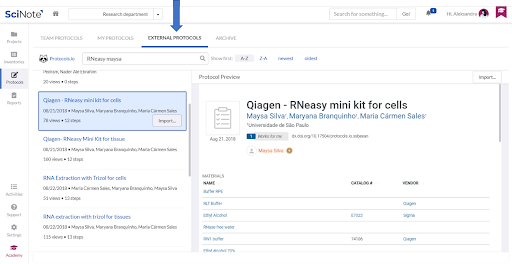
LOADING THE PROTOCOL FROM THE PROTOCOLS REPOSITORY ON THE NUCLEIC ACID EXTRACTION TASK
We simply loaded the imported protocol on the Nucleic Acid Extraction task (by the Load Protocol button). All the protocol steps were nicely arranged and we could edit them to add some details, add attachments or comments.
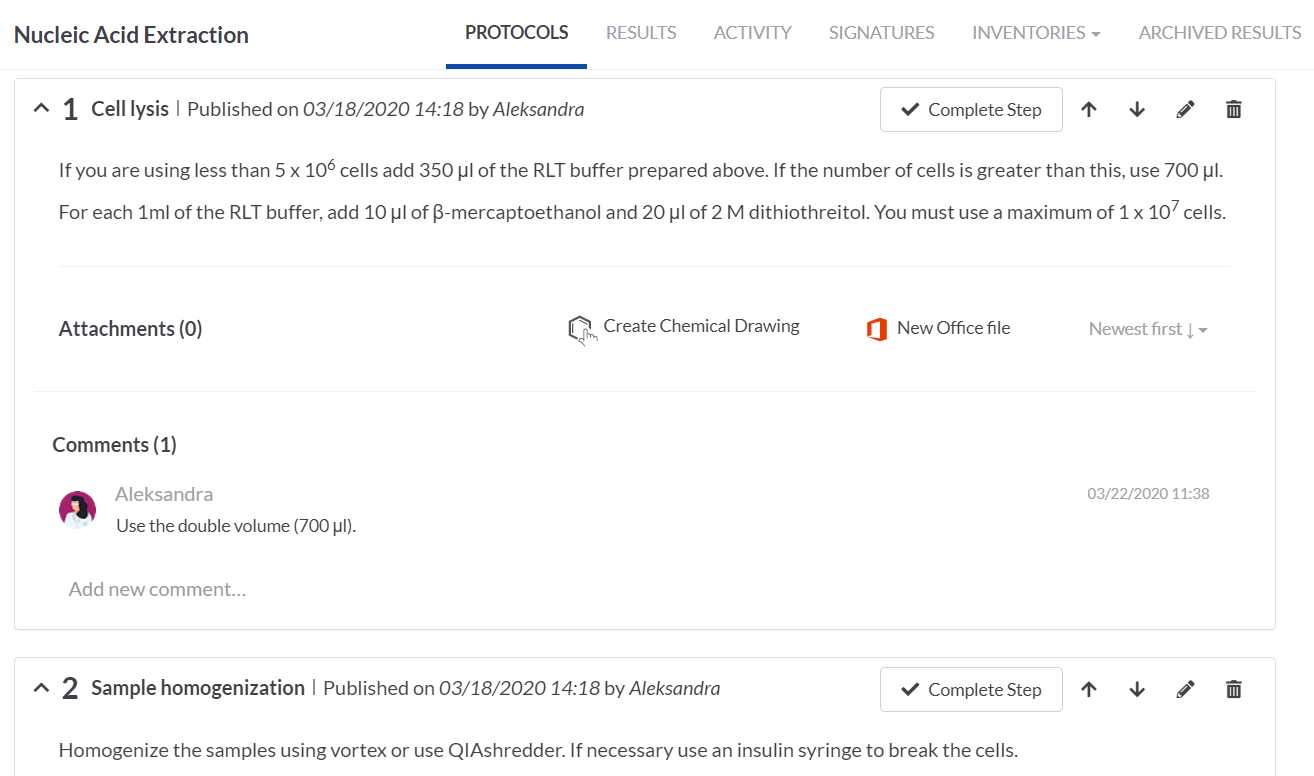
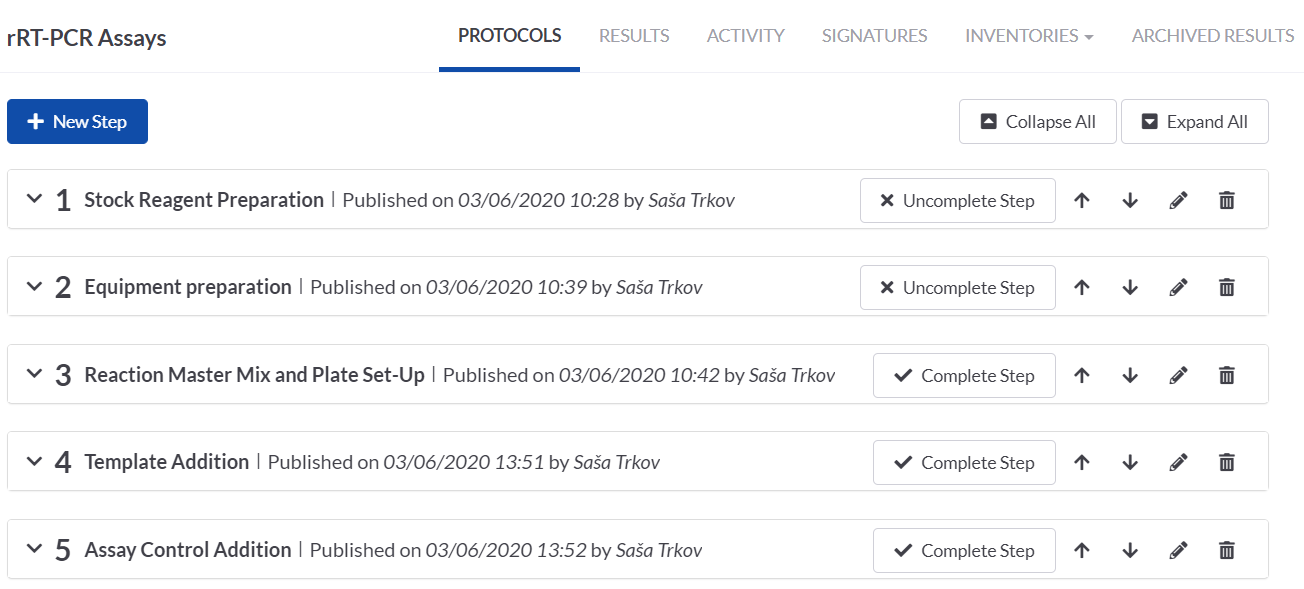
SETTING UP THE PROTOCOL STEPS ON THE rRT-PCR TASK
We then set up the protocol steps on the rRT-PCT task. The collapsed view of the steps gives a nice overview of the process and it can easily be tracked which steps have been completed and what follows.
ASSIGNING REAGENTS FROM THE INVENTORY ON THE TASK
We assigned the RT-PCR reagents from our inventory directly on the rRT-PCR task. This way, we can have a quick overview of the used reagents and their proper information, such as the manufacturer, catalogue number, storage conditions and the location, expiration date, data sheet files, etc. Assigning of the inventory items to the task can be done in the Inventories tab on the task.

ADDING THE TABLE (WITH CALCULATIONS) ON THE PROTOCOL STEP
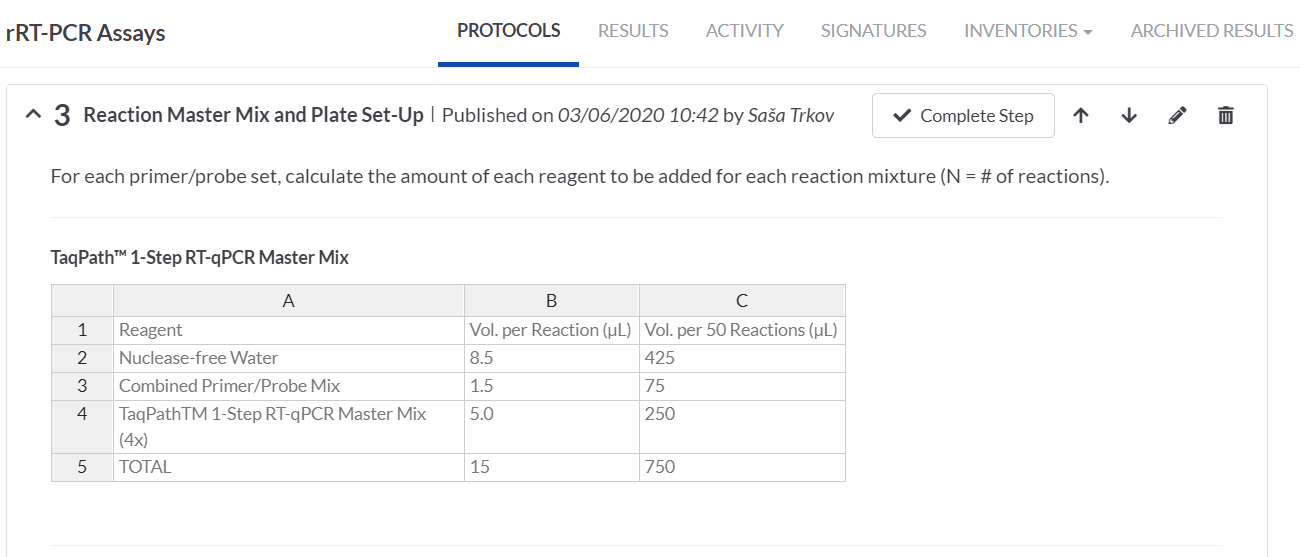
In the RT-PCR Assay protocol one of the steps is to prepare a reaction master mix and a plate set-up. For these we made use of the SciNote tables that allow calculations of the required volumes of the master mix components based on the number of our samples. Alternatively, we could also make the calculations in an Excel table and attach it on the step.
INSERTING THE ANNOTATED IMAGE ON THE PROTOCOL STEP
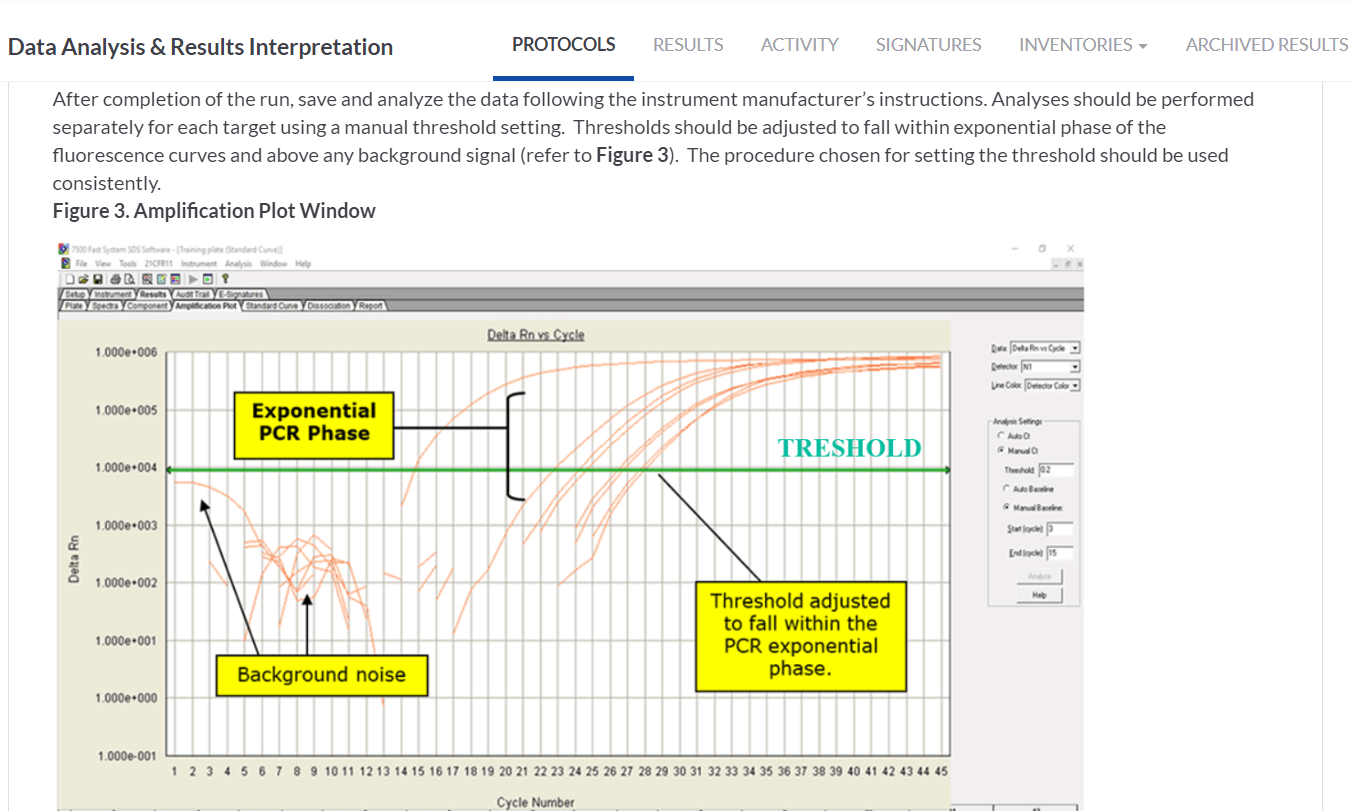
In the Data Analysis & Results Interpretation task we attached the representative image of the RT-PCR amplification plot with the instructions on how to read the output data from the published protocol. We made use of the annotation tools for the images attached on the step in SciNote to make some comments, explanations or emphasis on the image itself.
So, this is how we set-up the protocol for the detection of 2019-Novel Coronavirus by real-time RT PCR by using and explaining some of the SciNote functionalities. We can assure you that you can easily create a well-structured protocol in SciNote for this kind of study or any other that your research team is working on. We support all your research efforts and wish you good luck with your work!
By Sasa Trkov Bobnar, PhD, Application Specialist, SciNote